
Eda Yildirim, Ph.D. (University of California, Los Angeles)
Assistant Professor of Cell Biology
E-mail: eda.yildirim@duke.edu
412A Nanaline Duke Building, Box 3709
Duke University Medical Center
Durham, NC 27710
Telephone 919-668-0460
Fax 919-684-8090
Epigenetic regulation and nuclear organization in mammalian development and disease
The coding capacity of the genome is not controlled solely by DNA sequences. Rather, fine tuning is achieved through epigenetic factors that impact the accessibility of DNA sequences for enzymatic modifications, and through factors that participate in genome organization establishing a functional nuclear landscape. In the Yildirim lab, we study how epigenetic mechanisms, particularly those that are mediated by long noncoding RNAs (ncRNAs), complement gene expression, impact genome stability and define cell fate decisions. Other research in the lab focuses on understanding how spatial organization of the genome is achieved through interactions between chromatin and components of the nuclear envelope. We are interested in defining the molecular bases of these interactions and delineating their significance in driving gene expression and genome functions. We approach these two main areas of research by utilizing a variety of genetic, cell biological and genomics tools using mouse stem cells and mouse models. In the long run, a detailed understanding of the genetic, epigenetic, and cellular mechanisms that are mediated by long ncRNAs and spatial positioning will allow us to explore their causal roles in disease and cancer.
I. Long noncoding RNA mediated gene expression, gene stability and cell fate decisions
Noncoding RNAs (ncRNAs) are noncoding transcripts that take part in epigenetic mechanisms by providing RNA-directed silencing, aiding recruitment of chromatin modifying complexes and in some instances, presenting enhancer-like functions to boost transcription. Data from genome wide expression profiling studies of various human cancers have provided evidence for altered expression of long ncRNAs. However, how long ncRNA-mediated pathways may impact tumorigenesis is still poorly understood.
Mammalian X chromosome inactivation (XCI) provides a powerful model for studying the interplay between epigenetic mechanisms driven by ncRNAs and development. This female specific dosage compensation mechanism depends on expression of Xist long ncRNA, which coats and transcriptionally silences future inactive X (Xi) balancing X-linked gene expression between XX females and XY males (Fig 1). While Xist expression is crucial for initiation of XCI, its role in maintenance of XCI is not clear.
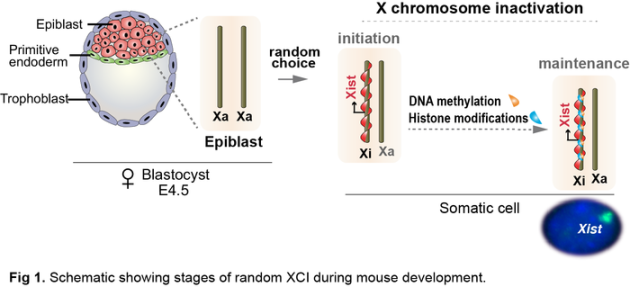
By deleting Xist specifically in blood cells of mice, we previously demonstrated that Xist plays an important role in hematopoietic stem cell (HSC) function and acts as a suppressor of cancer in mice (Yildirim et al., Cell 2013) suggesting a role for Xist in maintenance of XCI and a causal link between Xist loss and cancer.
Our laboratory uses primarily XCI as a model to understand how long ncRNAs 1) regulate gene dosage and maintain epigenetic state, 2) impact genome stability, and 3) participate in cell fate decisions. Our long-term goal is to delineate the molecular bases and order of genetic, epigenetic and cellular processes that link altered expression of long ncRNAs and pathways driven by these molecules to cancer.
II. Spatial organization of gene expression
Roles of nuclear envelope components in gene expression and genome organization
In addition to their canonical functions in maintaining nuclear architecture and nucleocytoplasmic trafficking, components of the nuclear envelope (i.e. nuclear pore complex (NPC), nuclear lamina) are also involved in intracellular signaling, DNA repair, and gene expression. In various organisms, components of the nuclear envelope interact with chromatin that is spatially organized within the nucleus (Fig 2) and subsequently regulate gene expression. Yet, the molecular mechanisms of these processes are poorly understood for mammalian systems.
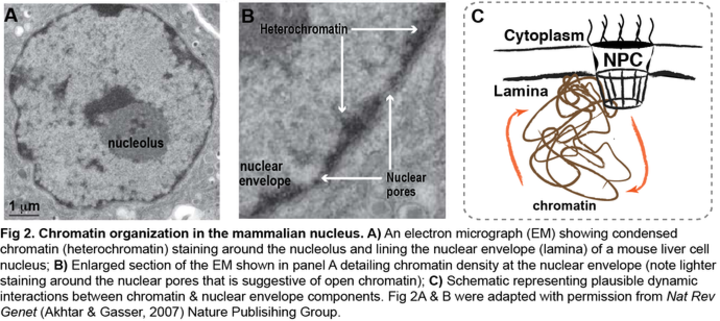
Our aim is to define the molecular bases of the interactions that are established between chromatin and the components of the nuclear envelope, and determine how they 1) regulate epigenetic state of genes and their transcription, 2) participate in spatial organization of the genome, and 3) contribute to cellular functions during mammalian development. Our findings will ultimately allow us to uncover how alterations in nuclear envelope components and chromatin configurations might induce uncontrolled cell divisions and migration patterns that lead to disease and cancer.
SELECTED PUBLICATIONS:
Yang, T., Ou, J. & Yildirim, E. Xist exerts gene-specific silencing during XCI maintenance and impacts lineage-specific cell differentiation and proliferation during hematopoiesis. Nat Commun 13, 4464 (2022).
Kadota S, Ou J, Shi Y, Lee JT, Sun J, Yildirim E. Nucleoporin 153 links nuclear pore complex to chromatin architecture by mediating CTCF and cohesin binding. Nat Commun. 2020 May 25;11(1):2606. doi: 10.1038/s41467-020-16394-3. PubMed PMID: 32451376; PubMed Central PMCID: PMC7248104.
Yang L, Yildirim E, Kirby JE, Press W, Lee JT. Widespread organ tolerance to Xist loss and X reactivation except under chronic stress in the gut. Proc Natl Acad Sci U S A. 2020 Feb 25;117(8):4262-4272. doi: 10.1073/pnas.1917203117. Epub 2020 Feb 10. PubMed PMID: 32041873; PubMed Central PMCID: PMC7049159.
Sun J, Shi Y, Yildirim E. The Nuclear Pore Complex in Cell Type-Specific Chromatin Structure and Gene Regulation. Trends Genet. 2019 Aug;35(8):579-588. doi: 10.1016/j.tig.2019.05.006. Epub 2019 Jun 15. Review. PubMed PMID: 31213386.
Yang, T., Shi, Y., Yildirim, E. (2017). Implications of Long Noncoding RNAs in Cancer Epigenetics. In “Cancer and Noncoding RNAs”. Translational Epigenetics Series. 1, 381-406. (Chakrabarti, J. and Sanga, M. eds), Elsevier Academic Press.
Yang, T., and Yildirim E. (2017). Epigenetic and LncRNA-mediated Regulation of X-chromosome Inactivation and Its Impact on Pathogenesis. Curr Pathobiol Rep. 10:1-12.
Pinter, S.F., Colognori, D., Beliveau, B.J., Sadreyev, R.I., Payer, B., Yildirim, E., Wu, C.T., and Lee J.T. (2015). Allelic imbalance is a prevalent and tissue-specific feature of the mouse transcriptome. Genetics. 200:537-549.
Yildirim, E., Kirby, J.E., Brown, D.E., Mercier, F.E., Sadreyev, R.I., Scadden, D.T., and Lee, J.T. (2013). Xist RNA is a potent suppressor of hematologic cancer in mice. Cell. 152, 727-742.
Sadreyev, R.I., Yildirim, E., Pinter, S.F., and Lee, J.T. (2013). Bimodial quantitative relationships between histone modifications for X-linked and autosomal loci. Proc Natl Acad Sci USA. 110, 6949-6954.
Yildirim, E., Sadreyev, R.I., Pinter, S.F., and Lee, J.T. (2012). X-chromosome hyperactivation in mammals via nonlinear relationships between chromatin states and transcription. Nat Struct Mol Bio. 19(1), 56-61. (First and second author equal contribution).
Pinter, S.F., Sadreyev, R.I., Yildirim, E., Jeon, Y., Ohsumi, T., Borowsky, M., and Lee, J.T. (2012). Spreading of X chromosome inactivation via a hierarchy of defined Polycomb stations. Genome Res. 10, 1864-76.