Science Highlights
Yan, et al. "An enhancer-based gene therapy strategy for spatiotemporal control of cargoes during tissue repair." Cell Stem Cell (2022). Read the article: https://doi.org/10.1016/j.stem.2022.1...
Xist exerts gene-specific silencing during XCI maintenance and impacts lineage-specific cell differentiation and proliferation during hematopoiesis
X chromosome inactivation (XCI) is a dosage compensation phenomenon that occurs in females. Initiation of XCI depends on Xist RNA, which triggers silencing of one of the two X chromosomes, except for XCI escape genes that continue to be biallelically expressed. In the soma XCI is stably maintained with continuous Xist expression. How Xist impacts XCI maintenance remains an open question. An exciting study by the Eda Yildirim Lab sheds light onto this question. Yang et al. investigated the functional and mechanistic roles of Xist in hematopoietic stem cell and progenitor cells (HSPCs) using the HSC-specific Xist knockout mouse model. They show that Xist deletion leads to lineage-specific differentiation defects and cell cycle alternations in HSPCs. By utilizing female HSPCs and mouse embryonic fibroblasts, they find that Xist loss leads to preferential upregulation of XCI escape genes compared to XCI subjective genes. Their mechanistic studies further reveal that Xist exerts gene-specific regulatory roles in controlling repression of transcription on the inactive X chromosome during XCI maintenance, which most likely underlie the variability in Xist-deficiency phenotypes between different cell lineages and tissues. These findings provide insights for treatment strategies of X-linked diseases through manipulating expression of specific X-linked genes on the inactive X chromosome.


Chemico-genetic discovery of astrocytic control of inhibition in vivo

In a collaborative project, the Soderling and Eroglu labs designed a new split-surface BioID approach to discover the proteomes mediating cell-cell interactions between neurons and astrocytes and how astrocytes control inhibitory GABAergic synapse formation and function in vivo. Their work is supported by a Co-PI NIH BRAIN initiative award. Read More
Mini-Lungs in a Lab Dish Mimic Early COVID-19 Infection -
Research team led by Purushothama Rao Tata
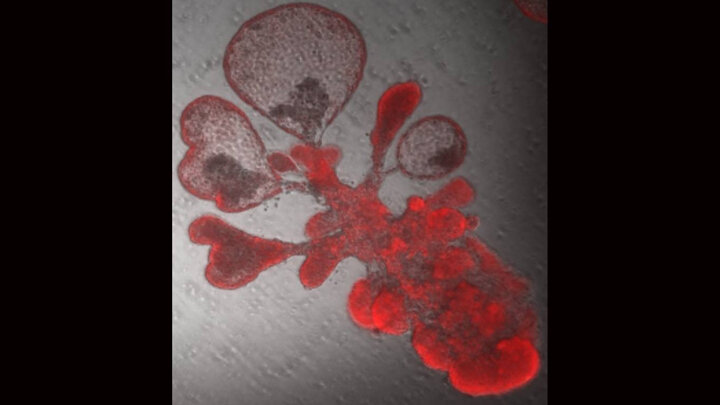
Lung Stem Cells in transit- a regenerative intermediate and a culprit in lung fibrosis.

Yoshihiko Kobayashi and Aleksandra Tata in the Tata Lab published a co-first author paper in Nature Cell Biology.
Gas exchanging regions of the lungs are covered by a very thin, flat and large cells known as alveolar type-1 cells (AT1). Upon injury, these damaged cells are replaced by a specialized stem cell population, also known as alveolar type-2 cells (AT2). However, how these small stem cells can generate such thin, flat and large AT1 cells remain unknown. In a recent published study, Tata lab used “mini lungs” (also known as organoids), multiple in vivo repair models, single-cell transcriptomics, and lineage tracing, we find that alveolar type-2 epithelial cells undergoing differentiation into type-1 cells acquire pre-alveolar type-1 transitional cell state (PATS) en route to terminal maturation. Transitional cells undergo extensive stretching during differentiation, making them vulnerable to DNA damage. PATS show an enrichment of TP53, TGFβ, DNA-damage response signalling, and cellular senescence. Gain and loss of function and genomic binding assays revealed a direct transcriptional control of PATS by TP53 signalling. Notably, PATS-like cells were accumulated in human fibrotic lungs, suggesting persistence of the transitional state in fibrosis. Thus, this study implicates a transient state associated with senescence in normal epithelial tissue repair and its abnormal persistence in disease conditions. Read More
Temperature-dependent sex determination is mediated by pSTAT3 repression of Kdm6b.

Ceri Weber, recent graduate in the Capel Lab, had an article published in Science.
In many reptiles, including the red-eared slider turtle Trachemys scripta elegans (T. scripta), sex is determined by ambient temperature during embryogenesis. We previously showed that the epigenetic regulator Kdm6b is elevated at the male-producing temperature and essential to activate the male pathway. Here, we establish a causal link between temperature and transcriptional regulation of Kdm6b. We show that STAT3 is phosphorylated at the warmer, female-producing temperature, binds the Kdm6b locus, and represses Kdm6b transcription, thereby blocking the male pathway. Influx of Ca2+, a mediator of STAT3 phosphorylation, is elevated at the female temperature and acts as a temperature-sensitive regulator of STAT3 activation.
Self-organized nuclear positioning synchronizes the cell cycle in Drosophila embryos.
Cell Paper from Di Talia Lab.
Synchronization of the cell cycle in syncytial Drosophila embryos is controlled by a self-organized mechanism that drives accurate nuclear positioning by integrating localized cell cycle oscillations, actomyosin cortical contractility, and cytoplasmic flows.
Plug and Play Protein Modification using Homology-independent Universal Genome Engineering. Neuron (2019).

Yudong Gao, PhD, from the Soderling Lab, developed a new method to make it much easier and faster for neuroscientists to study individual proteins in a living brain. Based on CRISPR gene editing technology, “Homology-independent Universal Genome Engineering”, or HiUGE, uses adeno-associated viruses to deliver multiple “plug and play” gene sequences to a variety of cells in a lab dish or a living organism. The new technique should revolutionize studies to identify, label and interpret the functions of individual proteins in higher throughput. (Neuron, July 1, 2019) Neuron PubMed