Chantell Evans, PhD (University of Wisconsin – Madison)
Assistant Professor of Cell Biology
HHMI Hanna Gray Fellow
Duke Science and Technology Scholar
E-mail: chantell.evans@duke.edu
Twitter: @channyskye @EvansLab
368B Nanaline Duke Building, Box 3709
Duke University School of Medicine
Durham, NC 27710
Mitochondrial surveillance in neuronal homeostasis and neurodegenerative disease
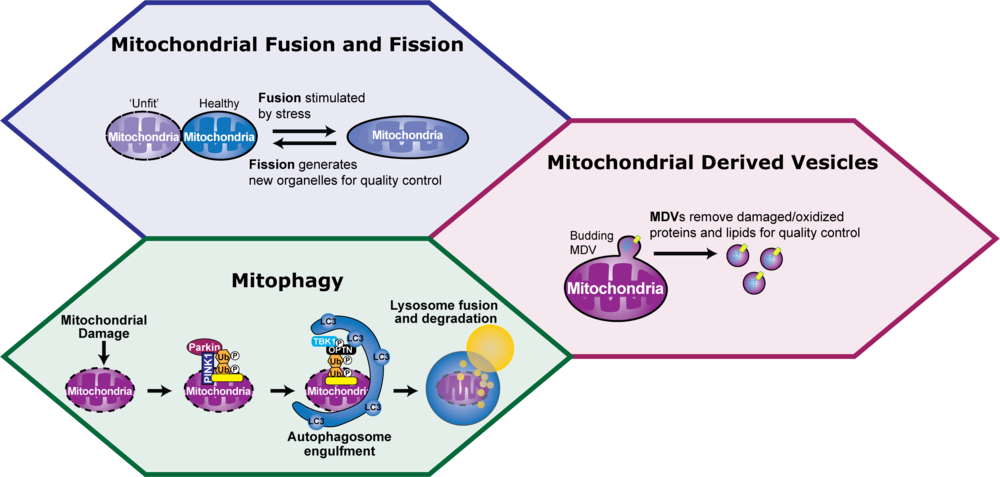
Neurons are post-mitotic, highly-polarized cells that must survive the lifespan of a human. They rely heavily on mitochondria to support local energy demands at distant fusion sites. These demands are supported by the continuous replenishment and maintenance of mitochondria, while aged or damaged mitochondria are removed from the cell via mitophagy. It is thought that mitochondrial dysregulation contributes to neurodegenerative diseases, such as Parkinson’s disease, Amyotrophic Lateral Sclerosis (ALS), and Alzheimer’s disease.
The complex morphology of neurons poses a unique challenge to maintain the mitochondrial network. A single human neuron is estimated to have two million mitochondria, where many organelles are located in far-reaching axons. How does a neuron survey every mitochondrion to continuously sustain a healthy mitochondrial population for the lifetime of a human? Emerging evidence suggests that multiple levels of mitochondrial surveillance are required, ranging from focal removal of damaged proteins and lipids to full degradation of dysfunctional organelles.
The focus of the Evans lab is to mechanistically define these diverse mitochondrial quality control pathways, including mitochondrial fusion and fission events, mitochondrial derived vesicles, and mitophagy. Our goal is to understand how these pathways collaborate to regulate the mitochondrial network in healthy neurons and what goes wrong in neurodegenerative disease. We integrate advanced live-cell microscopy to visualize mitochondrial dynamics with biochemical techniques and proteomics to define the specificity of these mechanisms in primary neurons, iPSCs, and mouse models of disease.
We currently have postdoctoral and technician positions available in the lab for highly motivated and talented applicants with interests in mitochondrial biology, neurodegenerative diseases, and quantitative imaging. Experience with mouse husbandry and handling, cell culture, or microscopy is preferred. Interested candidates should email a cover letter and CV to csevans@pennmedicine.upenn.edu.
Selected Publications:
Evans CS, Holzbaur ELF. Lysosomal degradation of depolarized mitochondria is rate-limiting in OPTN-dependent neuronal mitophagy. Autophagy. 2020 May;16(5):962-964. doi: 10.1080/15548627.2020.1734330. Epub 2020 Mar 4. PubMed PMID: 32131674.
Evans CS, Holzbaur ELF. Degradation of engulfed mitochondria is rate-limiting in Optineurin-mediated mitophagy in neurons. Elife. 2020 Jan 14;9. doi: 10.7554/eLife.50260. PubMed PMID: 31934852.
Evans CS, Holzbaur ELF. Quality Control in Neurons: Mitophagy and Other Selective Autophagy Mechanisms. J Mol Biol. 2020 Jan 3;432(1):240-260. doi: 10.1016/j.jmb.2019.06.031. Epub 2019 Jul 8. Review. PubMed PMID: 31295455.
Hoshino A, Wang WJ, Wada S, McDermott-Roe C, Evans CS, Gosis B, Morley MP, Rathi KS, Li J, Li K, Yang S, McManus MJ, Bowman C, Potluri P, Levin M, Damrauer S, Wallace DC, Holzbaur ELF, Arany Z.The ADP/ATP translocase drives mitophagy independent of nucleotide exchange. Nature. 2019 Nov;575(7782):375-379. doi: 10.1038/s41586-019-1667-4. Epub 2019 Oct 16. PubMed PMID: 31618756.
Evans CS, Holzbaur ELF. Autophagy and mitophagy in ALS. Neurobiol Dis. 2019 Feb;122:35-40. doi: 10.1016/j.nbd.2018.07.005. Epub 2018 Jul 5. Review. PubMed PMID: 29981842.